By deactivating a natural inhibitor of fatty acid synthesis, BIAM researchers have succeeded in increasing the oil content of microalgae fivefold. This is a major advance in the optimisation of bioenergy, paving the way for more efficient, sustainable production.
Fatty acids play an essential role in the structure of cell membranes, carbon storage and signalling. Using Crispr genome-editing tools (see inset), BIAM researchers have demonstrated that it is possible for microalgae to produce five times more fatty acids by deactivating a natural regulator that limits their production. A better understanding of how fatty acid production is regulated in the chloroplast (the small factory in plant cells where photosynthesis takes place) opens the way to new strategies for optimising this production in microalgae.
A first stage in the synthesis of fatty acids subject to multi-faceted regulation
It is well known that photosynthesis, with the help of light energy, transforms carbon from atmospheric CO2 into organic carbon and chemical energy. Cells then have to distribute their resources correctly to produce the various components essential to their functioning, such as proteins, sugars and fatty acids. An imbalance in this distribution, whether a deficit or an excess, could compromise the cell’s ability and survival.
The first stage in the de novo1 synthesis of fatty acids in the chloroplast relies on the action of a key enzyme in this process, acetyl-CoA carboxylase (ACCase). However, its activity is not constant: it is finely regulated by various signals that activate or inhibit it, making it possible to fine-tune the use of carbon and energy in fatty acid synthesis.
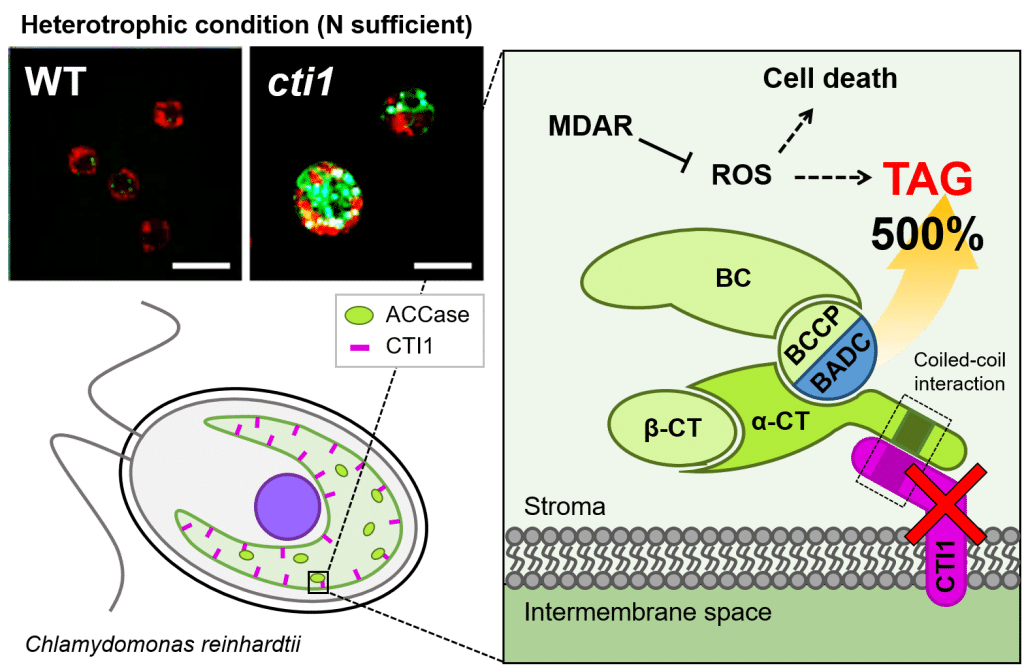
The inactivation of CTI1, a negative regulator of fatty acid synthesis, resulted in a five-fold increase in cellular oil content.
Identification of a key negative regulator of de novo fatty acid synthesis and its use to increase lipid content
As part of an international collaboration, BIAM researchers have discovered in Chlamydomonas a protein similar to Carboxyltransferase Interactor 1 (CTI1), already identified in other species. Carboxyltransferase is an essential enzyme in the manufacture of fatty acids, as it is involved in the early stages of the process. The homologue identified, named CrCTI1, interacts with a subunit of the ACCase complex and acts as a natural brake on fatty acid production.
Using advanced genome-editing tools, the team has demonstrated that deleting the CrCTI1 gene in algal cells results in a five-fold increase in oil content, without altering cell growth.
Unlike plants, where lipid accumulation depends mainly on light, the oil phenotype of crcti1 mutants is mainly influenced by trophic growth conditions. By growing the cells under heterotrophic conditions (where they feed on external carbon sources rather than light), the scientists observed a crucial function for CrCTI1 in regulating the balance between lipid accumulation and cell growth. Cellular analyses show that CrCTI1 plays an essential role in carbon management. It enables cells to adapt to the quantity of carbon available by controlling carbon metabolism, antioxidants and fat oxidation.
These results are a continuation of research into the regulation of lipid metabolism and strengthen the prospects for biotechnological applications. By gaining a deeper understanding of the mechanisms controlling the synthesis of fatty acids, it is now possible to quintuple their production, thereby increasing their potential as a source of renewable energy.
Genome editing
CRISPR genome-editing tools are like ultra-precise ‘genetic scissors ’i. They can modify an organism’s DNA by cutting a specific sequence to delete, add or correct it. It should be noted that these targeted adjustments are based on natural mechanisms already present in cells, offering unprecedented control over genome editing.
1 De novo fatty acid synthesis refers to the process by which cells make fatty acids from small molecules such as acetyl-CoA.
Authors :
Zhongze Li1, Minjae Kim1, 2, Jose Roberto da Silva Nascimento3, Bertrand Legeret1, Gabriel Lemes Jorge3, Marie Bertrand1, Fred Beisson1, Jay J Thelen3*, Yonghua Li-Beisson1*
Collaboration :
1Aix-Marseille Université, CEA, CNRS, Institute of Biosciences and Biotechnologies of Aix-Marseille, UMR 7265, CEA Cadarache, Saint-Paul-lez Durance F-13108, France
2Library of Marine Samples, Korea Institute of Ocean Science & Technology, Geoje 53201, Republic of Korea
3Department of Biochemistry and Interdisciplinary Plant Group, University of Missouri, Christopher S. Bond Life Sciences Center, Columbia, Missouri 65211, United States
References
Plant Biotechnology Journal. DOI 10.1111/pbi.14581