Microalgae and biomass
BIAM scientists have identified the crucial role of a plant-specific kinase in regulating daughter cell hatching in microalgae. This breakthrough enables better management of cell wall degradation, facilitating the release of daughter cells and biomass production.
One of the main challenges in using microalgae to produce fuel is to understand how to boost their lipid productivity. “The ability of microalgae to produce lipids depends both on the amount of lipids already present in the cell, and on the growth of the cell itself” explains Yonghua Li-Beisson, leader of the team behind the discovery. “In unicellular organisms such as microalgae, cell growth is linked to the reproductive cycle, which culminates in a decisive stage: cell hatching”.
The regulation of this process is indeed complex, and involves different cellular components. Plants and algae, for example, have different types of polysaccharides in their cell walls, but they share a common family of key molecules, the glycoproteins1. These molecules, made up of proteins and sugars, play a major role in cell structure and communication: “For cell growth and differentiation processes to proceed normally, the synthesis and degradation of these glycoproteins must be finely controlled. However, the precise mechanisms that regulate glycoprotein degradation in plants and algae are still poorly understood”, continues the scientist.
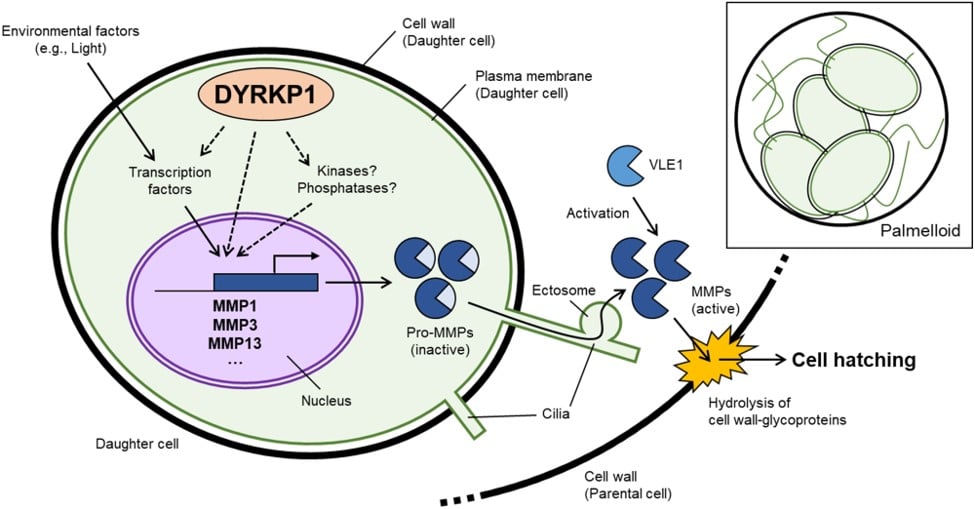
In the alga Chlamydomonas reinhardtii2, the cell cycle starts again after the hatching of daughter cells, a process regulated by a certain enzyme that plays a key role. These enzymes are matrix metalloproteinases (MMPs), which help to break down the cell wall to enable the release of daughter cells. “In previous research3 carried out by our institute, our teams highlighted an important player in this process: the DYRKP kinase, a plant-specific enzyme [P for plant]” she recalls.
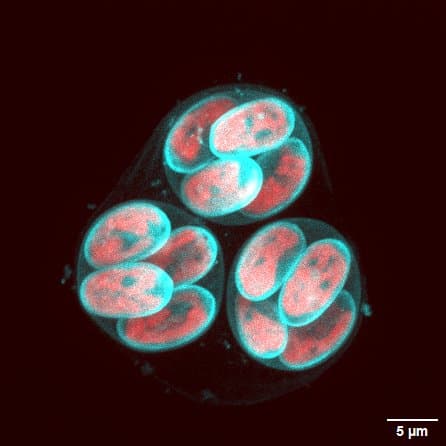
“In previous research3 carried out by our institute, our teams highlighted an important player in this process: the DYRKP kinase, a plant-specific enzyme [P for plant]” she recalls. “It was then revealed that DYRKP controlled MMP expression, and hence cell wall degradation. In the absence of this kinase, the cell wall cannot be properly broken down, trapping daughter cells inside the mother cell”, she explains. “Our latest study has just revealed that it is this dysfunction that leads to the formation of so-called palmelloid structures, trapping one inside the other.”
This discovery opens up a new avenue for improving lipid productivity in microalgae. Cell content and composition, particularly lipid content, depend on metabolic activity, which is in turn regulated by DYRKP. “DYRKP is a kinase specific to plants and algae. We are only just beginning to understand its actions. DYRKP is part of a larger family, called DYRK, which is found not only in yeast and plants, but also in bacteria and humans”, points out Yonghua Li-Beisson.
These results are crucial from both a fundamental and a practical point of view: they provide a better understanding of cell hatching, a key element in morphogenesis and cell growth. From the point of view of algal engineering, this knowledge should improve microalgae harvesting techniques, which are often costly.
1Glycoprotein: is a molecule composed of a protein attached to sugar chains. These molecules play an important role in many biological processes, such as cell-to-cell communication, cell recognition and cell protection. They are found on cell surfaces and in body fluids.
2Chlamydomonas reinhardtii : a microalga used as a model in plant biology.
3 Schulz-Raffelt et al 2016 https://pubmed.ncbi.nlm.nih.gov/26958078/
References
Minjae Kim1, Gabriel Lemes Jorge2, Moritz Aschern3,4, Stéphan Cuiné1, Marie Bertrand1, Malika Mekhalfi1, Jean-Luc Putaux5, Jae-Seong Yang3, Jay J. Thelen2, Fred Beisson1, Gilles Peltier1, and Yonghua Li-Beisson1
Collaboration:
2 Division of Biochemistry and Interdisciplinary Plant Group, Christopher Bond Life Sciences Center, University of Missouri; Columbia, Missouri, 65211, USA
3 Centre for Research in Agricultural Genomics (CRAG), CSIC-IRTA-UAB-UB, Campus UAB; Cerdanyola, 08193, Spain
4 Doctoral Program of Biotechnology, Faculty of Pharmacy and Food Sciences, Universitat de Barcelona; Barcelona, 08028, Spain
5 Grenoble Alpes University, CNRS, CERMAV; Grenoble, 38000, France